Lesson 3: What is Biowulf?
Bioinformatics Training and Education Program (BTEP)
Lesson 2 Review
- Directories are folders that can store files, other directories, links, executables, etc.
- The file system is hierarchical with the root directory (
/) at the top.
pwd= print working directory
ls= list contents
cd= change directory
mkdir,rmdir= make directory; remove directory
rm= remove file
touch= create file
nanoopens text editor- absolute file paths include the entire location from the root of the file system
- relative file paths include the location from the current working directory
Learning Objectives
- Understand the components of a HPC system. How does this compare to your local desktop?
- Understand when use of an HPC would benefit you.
- Learn how to connect to Biowulf
- Understand Biowulf data storage options
- Understand how to use the Biowulf module system
- Get help when needed; understand safeguards
Additional training materials at hpc.nih.gov
Content for this presentation is from hpc.nih.gov. For more information and more detailed training documentation, see hpc.nih.gov/training/.
NIH HPC Systems
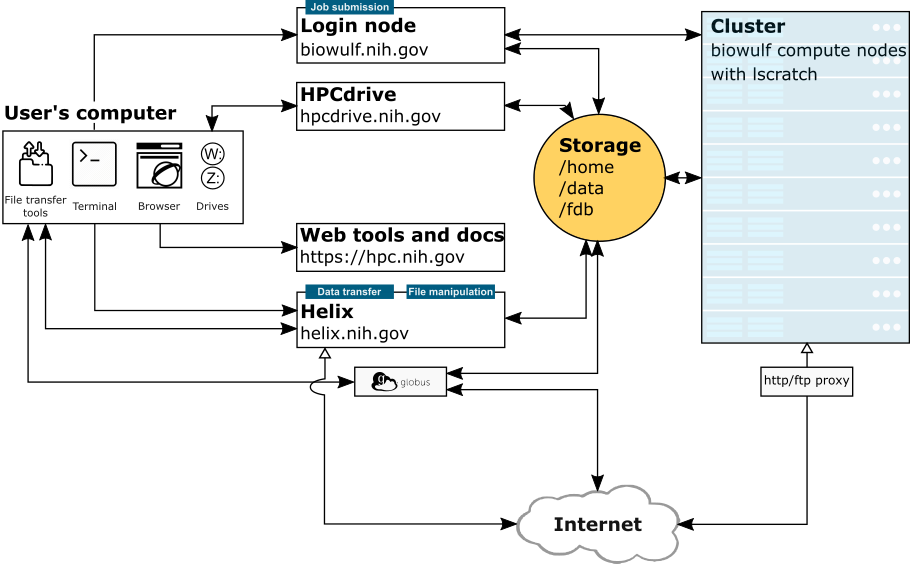
Image Credit: hpc.nih.gov/systems
What is Biowulf?
- The NIH high-performance compute cluster is known as “Biowulf”
- It is a 90,000+ processor Linux cluster
- Can perform large numbers of simultaneous jobs
- Jobs can be split among several nodes
- Scientific software (600+) and databases are already installed
- Can only be accessed on NIH campus or via VPN
Do not put data with PII (personally identifiable information), patient data for example, on Biowulf.
How does Biowulf compare to my local Desktop?
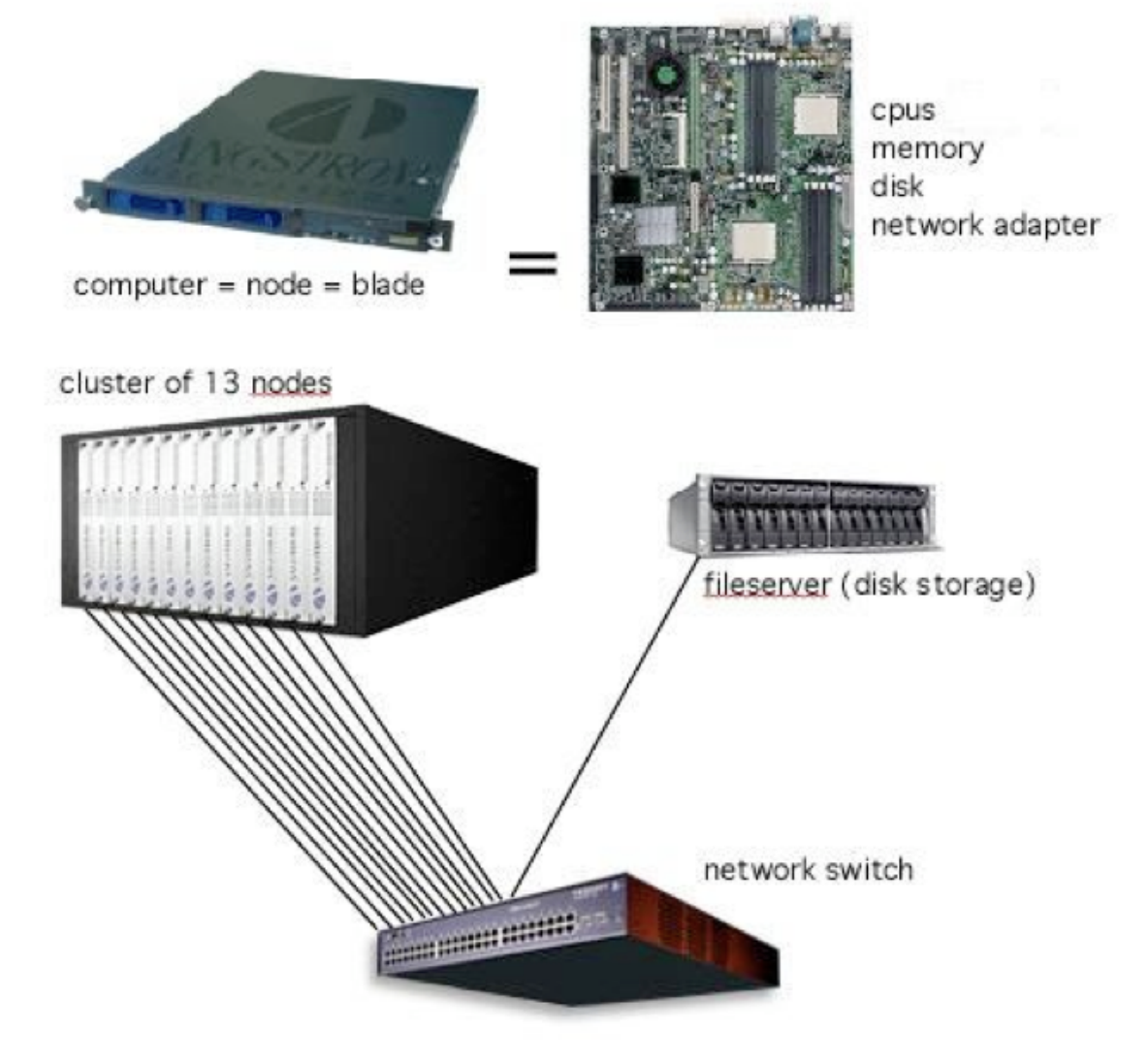
- Biowulf is a scaled up version of your local computer.
- Each compute node is like your individual computer; they have processors, memory, a disk drive, and a network adapter.
- In its simplest form, a cluster is simply several computers (nodes) clustered together, connected to a network switch, and some disk storage.
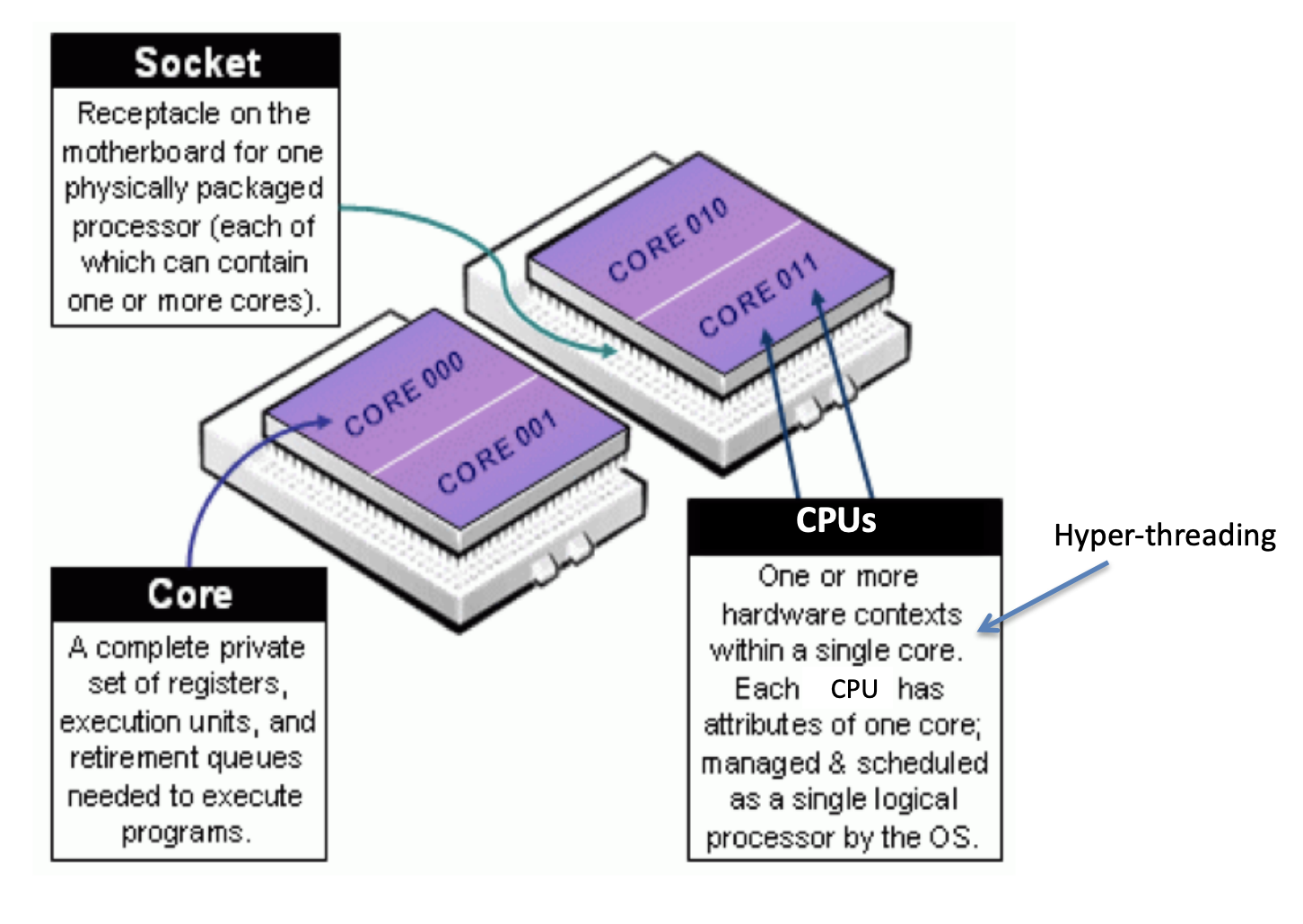
- Each node has two processors, which have multiple cores. Each core can support two threads of execution (via CPUs).
When should we use Biowulf?
- If software is unavailable or difficult to install on your local computer and is available on Biowulf.
- You are working with large amounts of data that can be parallelized to shorten computational time (e.g., Working with millions of sequences)
- Performing computational tasks that are memory intensive (e.g., whole genome assemblies)
- Working with large scale data
What is helix?
Helix is used for data transfers and file management on a large scale.
Transfering small files (< 1 GB) can be done from Helix, Biowulf login node, Globus, or hpc drive.
In general, Helix should be used when
- you are transferring >100 GB using
scp - gzipping a directory containing >5K files, or > 50 GB
- copying > 150 GB of data from one directory to another.
- uploading or downloading data from the cloud.
- you are transferring >100 GB using
For more information on data transfers see hpc.nih.gov.
Connecting to Biowulf
Connect remotely to Biowulf using a secure shell (SSH) protocol.
The login node will be used to submit jobs to run on the compute nodes that make up Biowulf.
ssh username@biowulf.nih.gov
“username” = NIH/Biowulf login username.
- If this is your first time logging into Biowulf, you will see a warning statement with a yes/no choice. Type “yes”.
- Type in your password at the prompt. NOTE: The cursor will not move as you type your password!
More on login nodes
Login nodes should be used for submitting jobs.
Other additional uses of login nodes include:
Editing/compiling code
File management (small scale)
File transfer (small scale)
Biowulf data storage
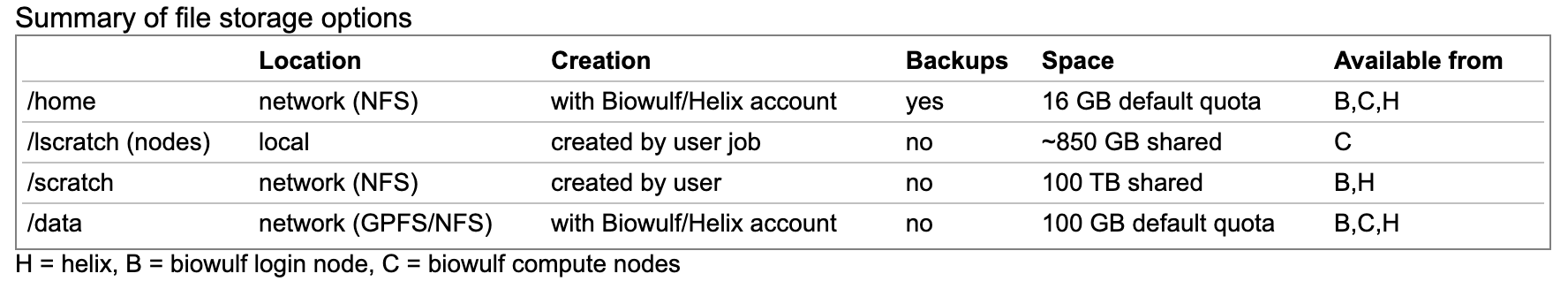
- You may request more space on /data, but this requires a legitimate justification.
Data storage on the HPC system should not be for archival purposes.
Check disk space
Use
checkquota - this shows the directories for which you have write access
OR
Look on the *user dashboard -> disk storage.
*Only works on VPN
What are snapshots?
A snapshot is a view of the directory at a specific point in time.
- Provides a copy of what existed at a given point in time.
- Useful for data recovery
BUT
- Are not back-ups.
Example of accessing a snapshot:
cd /home/$USER
ls -a
cd .snapshot
Best practices file storage
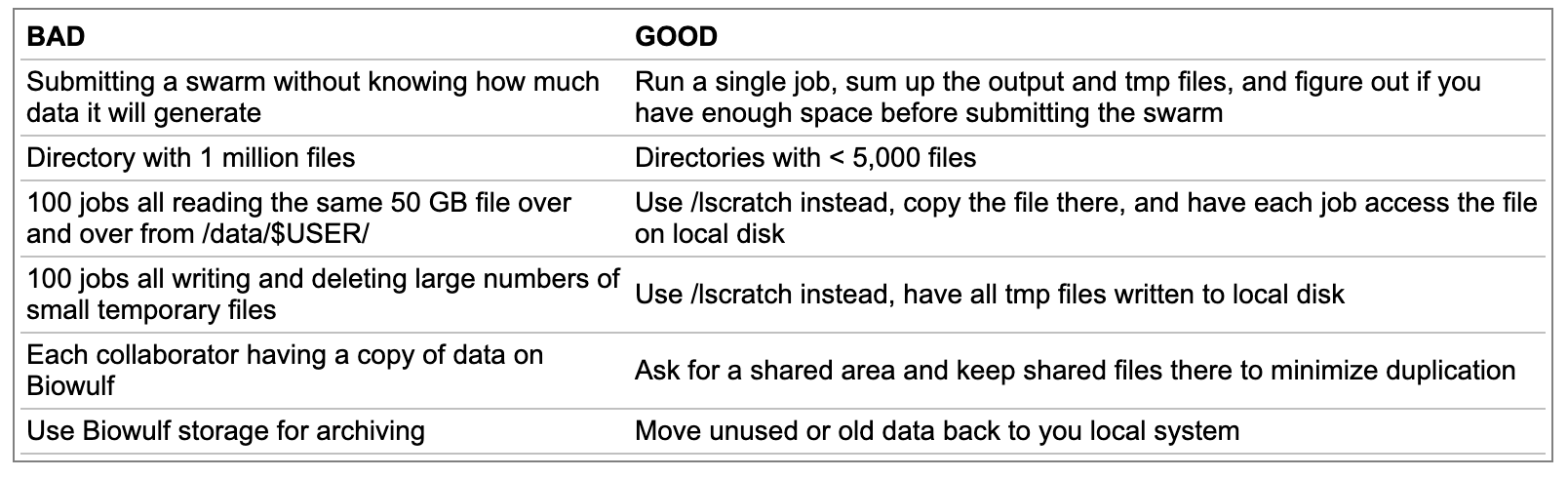
Create a directory in scratch:
mkdir /scratch/$USER
More on $USER and other environment variables
$USERis an example of an environment variable.
Environment variables contain user-specific or system-wide values that either reflect simple pieces of information (your username), or lists of useful locations on the file system. — Griffith Lab
We can display these variables using
echo.echo $USER
echo $HOME
$PATH
$PATHis an important environment variable.
echo $PATH
- This results in a colon separated list of directories containing programs that you can run without specifying those directories each time you run the program.
Adding to $PATH
You will likely need to add to your $PATH at some point in the future.
To do this use:
export PATH=$PATH:/path/to/folder
This change will not remain when you close the terminal. To permanently add a location to your path, add the above line to your bash shell configuration file, ~/.bashrc.
Biowulf module system
Many Bioinformatics programs are available on Biowulf via modules.
To see a list of available software in modules use
module avail
module avail [appname|string|regex]
module –d
How to load / unload a module
To load a module
module load appname
module load appname/versionTo see loaded modules
module listTo unload modules
module unload appname
module purge #(unload all modules)Note: you may also create and use your own modules
Biostars on Biowulf
There is a module on Biowulf called
biostars.
This module was created by the BTEP team and is not accessible in the Biowulf module system.
To load this module use:
module use /data/classes/BTEP/apps/modules module load biostarsSee more information in the course docs, Biostars on Biowulf
Batch system: slurm
- Most (almost all) jobs should be run in batch
- jobs can be submitted using
sbatchorswarm
- jobs can be submitted using
- Default compute allocation = 1 physical core = 2 CPUs in Slurm notation
- Default Memory Per CPU = 2 GB Therefore, default memory allocation = 4 GB
CPUs and memory should be designated at the time of job submission.
MORE ON THIS LATER
Getting Help
Existing safeguards make it nearly impossible for individual Biowulf users to irreparably mess up the system for others.
WORST CASE SCENARIO - You are locked out of your account pending consultation with NIH HPC staff
- NIH HPC staff want to help you!
- Contact them if you need help (staff@hpc.nih.gov)
- need help troubleshooting?
- need help with software installation?
- have a general concern?
- Attend monthly zoom-in consult sessions
- Contact them if you need help (staff@hpc.nih.gov)
- Check out user documentation
User Dashboard

- Can view disk usage and job info
- Request more disk space
- Evaluate job info for troubleshooting
Obtaining a Biowulf Account
- All NIH employees in the NIH Enterprise Directory (NED) are eligible for a Biowulf account.
- There is a charge of $35 per month associated with each account.
- Inactive accounts are locked after 60 days.
- You can unlock your account using your user dashboard.
- Apply for a Biowulf account here.
Summary
Biowulf is the high performance computing cluster at NIH.
When you apply for a Biowulf account you will be issued two primary storage spaces:
/home/$User(16 GB)
/data/$USER(100 GB).
Hundreds of pre-installed bioinformatics programs are available through the
modulesystem.
Computational tasks on Biowulf should be submitted as a job (
sbatch,swarm) or through an interactive session (sinteractive).
Do not run computational tasks on the login node.
Help Session
Task 1
Task 2
Task 3
Disk Storage, Checkquota, Snapshots