Join the NIDAP Bioinformatics Developer Community!
Our Mission
We are a group focused on using bioinformatics to advance cancer research. Working with us offers opportunities to collaborate with experts in the field. Interested? We’re eager to work together.
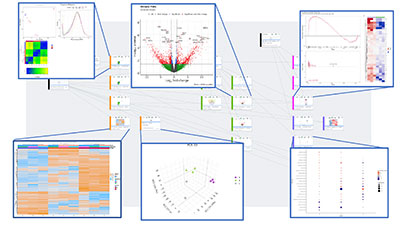
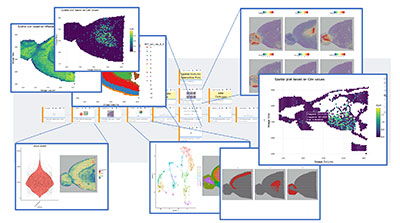
Active Workflows:
- Multipurpose Omics Workflow: a single flexible workflow that encompasses many different types of data
- Bulk RNA-Seq
- Proteomics
- Metabolomics
- miRSeq
- nCounter Gene Expression (Nanostring)
- Multimodal Single Cell Workflow
- RNA-Seq
- TCR-Seq
- CITE-Seq
- Spatial Transcriptomics Workflows
- Visium (10x Genomics) (Beta)
- DSP (Digital Spatial Profiling)
Benefits as a Developer:
By engaging with the NIDAP Developer Community, you’ll gain new skills and stay abreast of the latest advancements in bioinformatics workflows. This ensures that you remain at the forefront of your field, benefiting from the community’s shared wisdom and contributing to its growth.
Things you will learn:
- Advanced Techniques for Best Practices in the Analysis of Multiple Data Types
- Agile Development of Code Templates and Workflows in NIDAP (see video)
- Effective Strategies for Developing R Packages
- Utilizing GitHub for Development and Maintenance of Version Control
Current Developers: Core Group of Bioinformaticians who Develop on NIDAP
- LCBG (CCR)
- Aleksandra Michalowski
- CGR (DCEG)
- Chad Highfill
- Difei Wang
- Infrastructure Support (CBIIT)
- Rui He
- George Zaki
- LCBG (CCR)
- Aleksandra Michalowski
- CGR (DCEG)
- Chad Highfill
- Difei Wang
- Infrastructure Support (CBIIT)
- Rui He
- George Zaki
How to Join
E-Mail Us At: NCICCBRNIDAP@nih.gov
Meeting Times
- Thursdays @ 10AM
- Weekly Teams Discussions
- Fridays 2PM-5PM
- Developer Office Hours