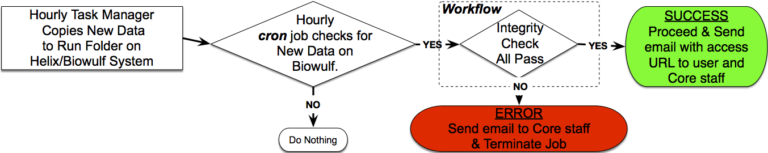
The standard starting point for the analysis of NextGen sequence data is the FASTQ file. These files contain not only the sequence information for each read, but also quality information about the accuracy of each base called. Unfortunately, Illumina’s NexSeq platform lacks an on-machine capacity to convert raw BCL files to standard FASTQ files. The need to address this problem, as encountered by CCR’s Genomics Core, induced us to develop a system that not only solved this problem, but also allowed the automated: generation of extensive QC data; the collection of per-run metadata; a simplified data archive and distribution scheme. Additionally, storing the metadata in a MySQL database, front-ended by a web-based interface has allowed us to easily generate usage reports and analytics across multiple sample runs. While this system was custom built for CCR’s Genomic Core, the basic principles could be applied to any NextSeq (or MiSeq) installation.
Detail of the administration web pages and QC reports can be found here
Design Goals
- Fully Automated System
- Supporting NextSeq & MiSeq Runs
- Converts Raw Data to Usable Sequence Files
- Easy Navigable QC Reports (numerous)
- Simple & Secure Distribution to Core-User
- Secure & Readily Accessible Archive
- Available Metadata — Web and MySQL
- Self-documenting & Attached Archive
- NCI’s Cleversafe Archive
- Core Facility Usage Reporting
- Initial self-signed URL generated by the main processing program – automatically sends email to staff and/or user
- The URL is valid for two weeks and can be used by a Web Browser or commands such as wget or curl to download the archive.
- New URLs can be generated at any time from the Web-based interface, or command line tools.
- URL’s are tied to READ-ONLY account for extra security.
- End user needs no special account, access privileges or tools.
- Object-Store system – flat name space, unique identifier, automatic encryption.
- Metadata can be directly associated with object.
- Robust – slices are distributed to separate disks and commodity hardware across geographic locations.
- Access via API calls from any system – follows Amazons S3 protocols
- Secure access via pre-signed URLs.
- Two NIH implementations – CIT (helix) and NCI Frederick
Data Retrieval & Email Notification
Cleversafe Presigned URL for Data Retrieval
- Cleversafe provides for URL based access to objects
- Workflow auto-generates URL and sends out emails
- Per NGS Run two packages are available for download
- Full data and QC Information — Gigabyte sized objects
- QC Information Only — Megabyte sized objects
- Secure access within NIH firewall with a two week time sensitive URL
- Re-issue URLs as needed by Core-facility
- Flexible download methods
- Use URL with any web browser and operating system
- MacOSX or Linux command line — curl or wget
Automated Email Generated and Sent to Designated Emails:
Instructs recipient on retrieval options along with a clickable hyperlink to their data + QC reports, or QC reports only. File sizes are provided as an indicator of the time required to download.
Sample Email
Dear Core User,
Your NextSeq data run on 170710 in data directory "170710_NB501558_0121_AHCYNGAFXX" with experiment name “Project Title Here” completed successfully
The data is accessible from the following URL:
http://cleversafetest.nci.nih.gov/SEQ37V/NEXTSEQ/170710_NB501558_0121_AHCYNGAFXX.zip?Signature=%2F%2BKeLNGRsRwAskpRbqXunM8p66w%3D&Expires=1503406626&AWSAccessKeyId=l6JcnKaAlMCbhfDQpECs
Download size is 18119603161 bytes (18.12 Gbytes).
############################# Notes on Obtaining your data #############################
There are two ways to obtain your data from an NIH Network attached computer:
1) Use the url link above.
2) Cut and paste the following command in a terminal window:
wget “http://cleversafetest.nci.nih.gov/SEQ37V/NEXTSEQ/170710_NB501558_0121_AHCYNGAFXX.zip?Signature=%2F%2BKeLNGRsRwAskpRbqXunM8p66w%3D&Expires=1503406626&AWSAccessKeyId=l6JcnKaAlMCbhfDQpECs” -O 170710_NB501558_0121_AHCYNGAFXX.zip
You have two weeks to retrieve your data. If the link expires, please contact Val Bliskovsky<bliskovv@mail.nih.gov>
If you wish to download only the QC Package. Use the following URL:
- Coded in: Python, C, shell scripts, mysql, php (web), javascript (web), plotly (web)
- Cron – C code – calling sbatch shell script
- bcl_process.py, bcl_move.py, miseq_move.py (python)
- modular structure for all variations
- bcl2fastq, fastqc, fastqscreen, multiQC
- transfer via boto3 (python calls) or gof3r
- Web interface (php, javascript, plotly, mysql database)