Lesson 1: Overview and logging on to Biowulf
Learning objectives
After this lesson, participants will be able to
- Provide reasons for learning Unix command line
- Describe Biowulf and why it is useful for NIH researchers
- Know how to obtain a Biowulf account
- Log on to Biowulf
- Find Biowulf help documentation
- Explore the Biowulf user dashboard
Commands that will be discussed
ssh: securely connect to remote computerpwd: to find present working directorycd: change into one directory from another
Reasons to use Biowulf
Biowulf is the Unix-based high performance computing system at the National Institutes of Health. Below are reasons for using Biowulf.
- Many bioinformatics programs/tools are written for the Unix operating system
- There 900+ programs/modules installed on Biowulf, including those used for Bioinformatics
- Current, past, and future versions of tools and databases available
- Reproducibility – written scripts/programs keep track of analyses steps
- Big data analysis – can open and work with very large data files
- Compute Power -
- has over 100,000 processor nodes
- large storage capacity (30+ petabytes)
- Globus for transfer of large data files
Skills learned while working on Biowulf apply to other high performance computing systems.
Warning
Do not store personally identifiable information on Biowulf!
Getting a Biowulf account
Information for obtaining a Biowulf account can be found at https://hpc.nih.gov/docs/accounts.html. The following conditions have to be met for Biowulf staff to grant accounts.
- PI approval
- PI pays $35 a month
- Annual renewal, which also requires PI approval
Biowulf student accounts
Each participant will be assigned a student account.
See here for student account assignment. Please use this account throughout this course. Enter NIH credentials to see the student account assignment sheet after clicking the link.
Connecting to Biowulf
To sign onto Biowulf, the users must be connected to the NIH network either by being on campus or through the VPN.
Windows 10 or above users will need to open the Command Prompt (type cmd in the Windows search box) while Mac users will need to open the Terminal application (type terminal in Spotlight search).
Once the Windows Command Prompt or Mac Terminal is opened, type the following to connect to Biowulf, where ssh is the command used to securely connect to a remote computer. Replace username with the assigned student account ID.
ssh username@biowulf.nih.gov
Note
For those who already have a Biowulf accounts or will obtain one in the future, use NIH username and password to connect.
Hit enter to supply the password.
The following message appears for those logging on to Biowulf for the first time. Respond with "yes" to proceed.
The authenticity of host 'biowulf.nih.gov (128.231.2.9)' can't be established. ECDSA key fingerprint is SHA256:BoP/KLS17g+gUuQ7mrCHa9oPPO+MHi/h8WML44iA1dw. Are you sure you want to continue connecting (yes/no)? yes
Once logged onto Biowulf, type the following to see the present working directory.
pwd
Users will start at the home directory upon signing onto Biowulf. Again, replace username with the student account ID for this class.
/home/username
The following command will take the user to the data directory. More on the home and data directories in Lesson 2.
cd /data/username
Biowulf website
The Biowulf webiste (https://hpc.nih.gov) has a menu that allows users to find useful information regarding the cluster (see Figure 1). Some of the useful tabs in this menu are discussed below.

Figure 1: Menu on the Biowulf website.
Applications
Need to find out what software are available on Biowulf? Then click on the Applications tab (Figure 2). The softwares are classified according to discipline.
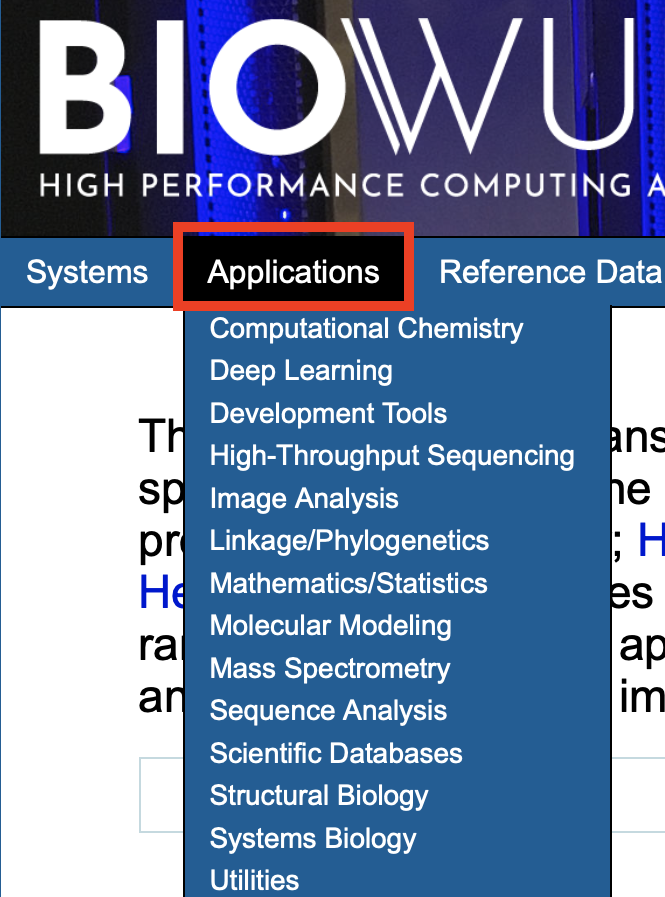
Figure 2:
Reference data
Analysis of NGS data requires reference genomes and annotations. Click on the Reference Data tab to see which are installed on Biowulf.
Training
Biowulf staff offer extensive trainings. To see what is available, click on the Training tab (Figure 3).

Figure 3: Find training offered by Biowulf staff
User dashboard
The User Dashboard provides
- Account information including group affiliations
- Disk usage and link to increase storage quota for user's data directory
- Information on submitted jobs
- Usage report
There is also a student dashboard for the student accounts.
Lesson 2 sneak peak
Below are the commands that will be introduced in Lesson 2.
ls: list directory contentchmod: change file and directory permissionspwd: get present working directorycd: change directorycp: copyrm: delete