Lesson 2: Biowulf directory structure
Quick review
Lesson 1 introduced the benefits of working on Biowulf and method for connecting to Biowulf from a personal computer by using the ssh command.
Learning objectives
After this lesson, participants should be able to
- Understand the Biowulf directory structure
- Describe the home and data directories on Biowulf
- Find help for Unix commands
- List directory content
- Describe file and folder permissions
- Change into one directory from another
- Copy files and folders
- Download files from the web
- Unpack tar files
- View file contents
Commands that will be discussed
ls: list directory contentchmod: change file and directory permissionspwd: get present working directorycd: change directorycp: copyzcat: to view compressed filescat: to view file content
Before getting started
Sign onto Biowulf using the assigned student account. Remember, Windows users will need to open the Command Prompt and Mac users will need to open the Terminal. Also remember to connect to the NIH network either by being on campus or through VPN before attempting to sign in. The command to sign in to Biowulf is below, where username should be replaced by the student ID.
ssh username@biowulf.nih.gov
See here for student account assignment. Enter NIH credentials to see the student account assignment sheet after clicking the link.
Biowulf directory path structure
The first step to navigating around Biowulf is to understand the directory path structure. The root folder is the top level folder in the Biowulf file system (Figure 1). In Unix systems, the root folder is designated by "/". Inside the root are the home and data directories. As an example, the data directory contains a folder P, which contains folders P_in and P_out (Figure 1).
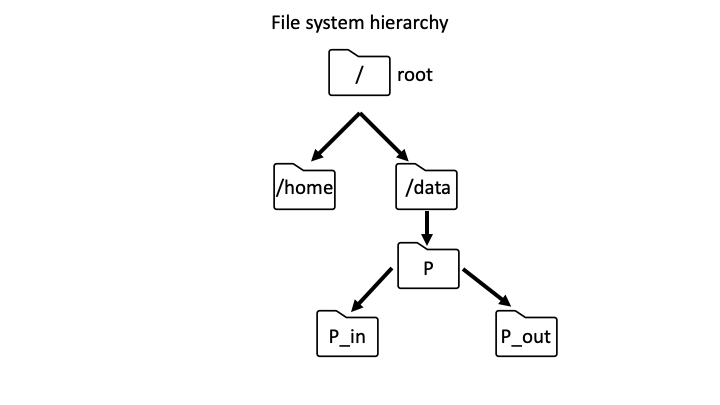
Figure 1: Example of Biowulf file system hierarchy.
Listing the contents of the root folder in Biowulf shows that the home and data directories reside within it. To list directory content, use the ls command followed by the name of the directory (in this case /, which denotes the root folder).
ls /
home
data
Home and data directories
Upon signing onto Biowulf, the prompt below appears (replace username with the assigned student ID). The ~ at the prompt denotes the home directory. The pwd command can be used to print the path to the present working directory, which should be home.
[username@biowulf ~]$
pwd
/home/username
The home directory is limited to 16 gb of storage space and cannot be expanded. Use the data directory, which has more default storage space and can be expanded when for data intensive analysis. . To change into the data directory use the cd command followed by the name of the folder (in this case it is /data/username). Replace username with the student account ID.
cd /data/username
The pwd command can confirm that change of directory was successful.
pwd
/data/username
Note
When pwd is used, the directory path retrieved starts at the "/" or the root. For instance, /home/username and /data/username. A directory path that starts from the root is known as an absolute path.
Finding help
To get help with Unix commands, use the man command, which pulls up the manual. Another option, which is command specific is to append either -h or --help to the command.
man man
man pwd
Hit q to exit the manual.
man ls
OR
ls --help
Viewing detailed directory content
Take a look at the /data/classes/BTEP folder using ls -l where the -l option lists directory contents in the detailed form
ls -l /data/classes/BTEP
Among the items in /data/classes/BTEP is a folder called unix_on_biowulf_2023_documents. The permission block (ie. the string drwxrwsr-x) for unix_on_biowulf_2023_documents begins with a "d", which denotes that it is a folder. A "-" at the beginning of block indicates a file. Figure 2 explains the permission block. File and folder permissions are important in Unix because it determines who can view and modify content.
drwxrwsr-x. 4 wuz8 GAU 4096 Feb 9 21:28 unix_on_biowulf_2023_documents
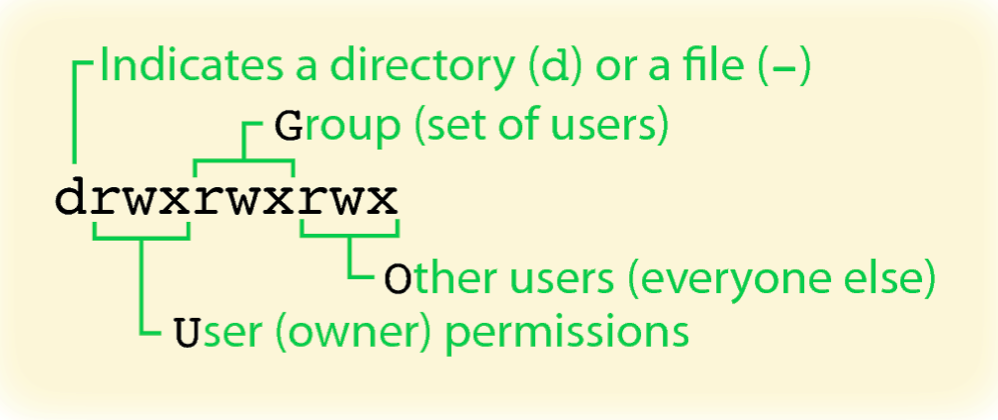
Figure 2: Unix permission block. The permissions are divided into three chunks of "rwx", corresponding to read, write, and execution privileges of the file or directory for owner, others in the group, and everyone else. If the permission begins with "d", then we are looking at a directory. If the permission begins with "-", then we are looking at file. Source: UF Research Computing.
The chmod command enables users to change file and folder permissions. To learn how to use this command use one of the following.
chmod --help
OR
man chmod
Copying a directory
For this part of the class, be sure to stay in the /data/username folder. If unsure, use pwd to check and use cd /data/username to change into if not in the folder.
Copy the unix_on_biowulf_2023_documents in /data/classes/BTEP to /data/username using the following cp command construct where the options and arguments are as follows.
- Option:
-rindicates to copy a folder-r: copy directories recursively
- Argument: name of of the folder to be copied (ie. /data/classes/BTEP/unix_on_biowulf_2023_documents)
- Argument: destination to copy the folder to (ie. /data/username, again, replace username with the assigned student ID)
cp -r /data/classes/BTEP/unix_on_biowulf_2023_documents /data/username
Note
The present working directory can be denoted by "."
Now, change into the unix_on_biowulf_2023_documents and look at the content using ls -l, which shows two files and two folders.
cd unix_on_biowulf_2023_documents
ls -l
-rwxr-x---. 1 wuz8 wuz8 368 Sep 5 11:26 SRP045416.swarm
drwxr-x---. 2 wuz8 wuz8 4096 Sep 5 11:26 SRR1553606
drwxr-x---. 2 wuz8 wuz8 4096 Sep 5 11:26 unix_on_biowulf_2023
-rwxr-x---. 1 wuz8 wuz8 41734 Sep 5 11:26 unix_on_biowulf_2023.zip
To go back to the /data/username folder (ie. one folder up) use cd with the .. notation.
cd ..
Copying a file
For this exercise, go back to the unix_on_biowulf_2023_documents folder in the data directory.
cd unix_on_biowulf_2023_documents
Make a copy of SRP045416.swarm and call it SRP045416_copy_1.swarm. To do this use the cp command where the arguments are
- File to make a copy of (ie. SRP045416.swarm)
- Name of the copy (ie. SRP045416_copy_1.swarm)
cp SRP045416.swarm SRP045416_copy_1.swarm
Go back to the data folder by doing
cd ..
Copy the SRP045416.swarm file in unix_on_biowulf_2023_documents here using the cp command where the arguments are
- File to copy (ie. SRP045416.swarm; here the relative path of unix_on_biowulf_2023_documents/SRP045416.swarm to the file is provided)
- Destination to copy the file (ie. "." which denotes here in the current directory)
cp unix_on_biowulf_2023_documents/SRP045416.swarm .
Note
Relative path is defined as the path related to the present working directory (pwd). It starts at your current directory and never starts with a / ." -- https://www.geeksforgeeks.org/absolute-relative-pathnames-unix/
Downloading from the web
Change back to the /data/username directory for this exercise. Replace username with the student account ID.
cd /data/username
There maybe times when it is necessary to download a data from the web. Use either wget or curl to download from the web. Here, curl will be shown where the options and arguments are
- Option:
-oto specify filename of the download - Argument: url for the file (ie. http://genomedata.org/rnaseq-tutorial/practical.tar). Note that the last part of the url (pratical.tar) is the filename but the
-ooption incurlenables saving of this file as something else.
curl -o hcc1395_fastq.tar http://genomedata.org/rnaseq-tutorial/practical.tar
List the directory content after download to confirm that hcc1395_fastq.tar is there.
Unpacking tar files
The hcc1395_fastq.tar is actually known as a tape archive (it has the .tar extension), which is a bundle of files and folders. The tar command is used to unpack its contents. The following are options and arguments used in the tar command to extract items.
- Options:
-x: extract files from an archive-v: verbosely list files processed-f: use archive file or device ARCHIVE- Argument: name of the file to unpack (ie. hcc1395_fastq.tar)
tar -xvf hcc1395_fastq.tar
The tar command should have unpacked the contents of hcc1395_fastq.tar into the /data/username directory. These are fastq files containing sequences derived from NGS experiment. These fastq files were compressed (.gz extension) to reduce storage space. Many bioinformatics algorithms can take fastq.gz as input so no need to uncompressed these.
hcc1395_normal_rep1_r1.fastq.gz
hcc1395_normal_rep1_r2.fastq.gz
hcc1395_normal_rep2_r1.fastq.gz
hcc1395_normal_rep2_r2.fastq.gz
hcc1395_normal_rep3_r1.fastq.gz
hcc1395_normal_rep3_r2.fastq.gz
hcc1395_tumor_rep1_r1.fastq.gz
hcc1395_tumor_rep1_r2.fastq.gz
hcc1395_tumor_rep2_r1.fastq.gz
hcc1395_tumor_rep2_r2.fastq.gz
hcc1395_tumor_rep3_r1.fastq.gz
hcc1395_tumor_rep3_r2.fastq.gz
Viewing file content
Stay in the /data/username folder and take a look at hcc1395_normal_rep1_r1.fastq.gz using the command zcat, which is used to view compressed files.
zcat hcc1395_normal_rep1_r1.fastq.gz
FASTQ file store the sequencing reads derived from Next Generation Sequencing. Each read is composed of four lines.
- Metadata header that starts with @
- The actual sequence
- "+"
- Quality score of each base in that read
@K00193:38:H3MYFBBXX:4:1101:10003:44458/1
TTCCTTATGAAACAGGAAGAGTCCCTGGGCCCAGGCCTGGCCCACGGTTGTCAAGGCACATCATTGCCAGCAAGCTGAAGCATACCAGCAGCCACAACCTAGATCTCATTCCCAACCCAAAGTTCTGACTTCTGTACAAACTCGTTTCCAG
+
AAFFFKKKKKKKKKKKKKKKKKKKKKKKKFKKFKKKKF<AAKKKKKKKKKKKKKKKKFKKKFKKKKKKKKKKKFKAFKKKKKKKKKKKKKKKKKKKKKKKKKKKFKKKKKKKKKKKKFKKKKKKKKKKKKFKFFKKKKKKKKKKKKFKKKK
Hit control c to exit zcat
Change into the unix_on_biowulf_2023_documents folder.
If pwd is /data/username then do the following. Remember to replace username with the student account ID.
cd unix_on_biowulf_2023_documents
If pwd is not /data/username then do the following
cd /data/username/unix_on_biowulf_2023_documents
To look at file content, use the cat command.
cat SRP045416.swarm
#SWARM --job-name SRP045416
#SWARM --sbatch "--mail-type=ALL --mail-user=wuz8@nih.gov"
#SWARM --gres=lscratch:15
#SWARM --module sratoolkit
fastq-dump --split-files -X 10000 SRR1553606
fastq-dump --split-files -X 10000 SRR1553416
fastq-dump --split-files -X 10000 SRR1553417
fastq-dump --split-files -X 10000 SRR1553418
fastq-dump --split-files -X 10000 SRR1553419
Note that cat can be used to view hcc1395_normal_rep1_r1.fastq.gz if it was uncompressed.
Change into the unix_on_biowulf_2023 folder.
cd unix_on_biowulf_2023
cat text_1.txt
oranges
blue
bananas
cats
dogs
apple
florida
gators
gainesville
alachua
county
btep