Recently, the NIH HPC Team has provided on demand access to HPC resources via web browser through integration of Open OnDemand, an open source project supporting “nearly 400 HPC centers around the world”.
This integration makes working with HPC resources less intimidating for new users, as they will not have to interact with command line applications to remotely connect via `ssh`. Instead, they can navigate to their web browser (Google Chrome is preferred) and connect to NIH HPC OnDemand using https://hpcondemand.nih.gov/. This requires access to the NIH network and authentication via PIV or MFA.
HPC OnDemand provides an online dashboard (See below) for users to easily access command line interactive sessions, graphical linux desktop environments, and interactive applications including RStudio, MATLAB, IGV, iDEP, VS Code, and Juptyer Notebook. This is not a separate service; it is a different interface for the HPC systems. As such, it uses the same account, resources, file systems, and job system as the rest of Biowulf. However, access is far easier than other methods (e.g., using Nomachine or SSH tunneling) and is recommended.
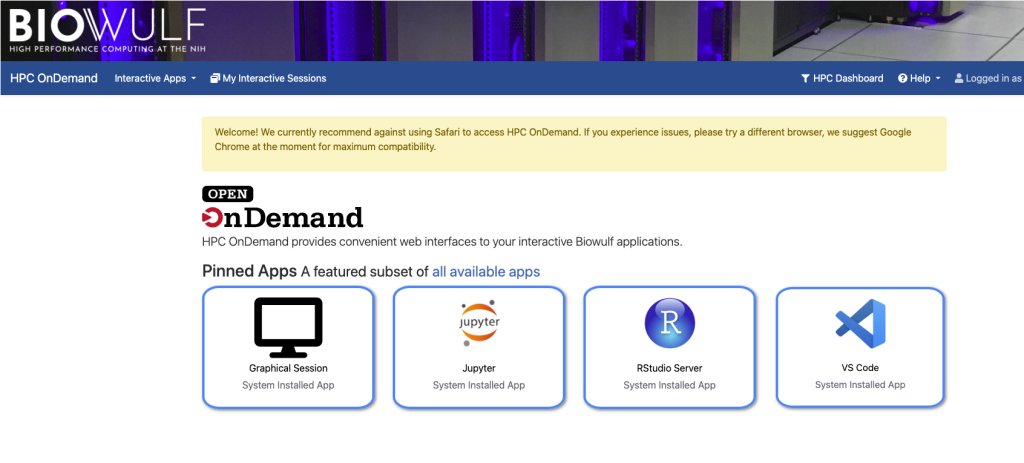
HPC OnDemand Dashboard: https://hpcondemand.nih.gov/
To use these interactive applications, Open OnDemand users will need to select computational resources (e.g., CPUs, memory, etc.) using a drop down menu, which means they should still have an idea about the resources needed to complete a given analysis.
If you are interested in learning more about HPC OnDemand on Biowulf, do not miss this month’s Coding Club on March 26th at 11 AM, “Getting Started with HPC OnDemand: A New Interface for NIH HPC Users”, which will introduce everything you need to know about the OnDemand platform and more.
Should you need additional help with HPC OnDemand or related issues, check out the resources below:
HPC OnDemand Documentation: HPC OnDemand is well documented on the NIH HPC website.
Contact staff@hpc.nih.gov: The HPC team welcomes questions and is happy to offer guidance to address your concerns.
Monthly consult sessions: The HPC team offers monthly consult sessions via Microsoft Teams. “All problems and concerns are welcome, from scripting problems to node allocation, to strategies for a particular project, to anything that is affecting your use of the HPC systems. The meeting connection details are emailed to all Biowulf users the week of the consult.”
Bioinformatics Training and Education Program: If you experience any difficulties or challenges, especially with different bioinformatics applications, please do not hesitate to email us at BTEP.
– Alex Emmons (BTEP)