High resolution single cell profiling assays have provided an unprecedented view of many biological systems and processes, but the spatial context in which this biology is occurring is often crucial. Spatial profiling, including spatial transcriptomic methods have emerged as a powerful way to study the spatial context of biological phenomena. Recent advancement in the sensitivity, scale and accessibility of these technologies are making their utilization in biomedical research more common. One popular platform for spatially-resolved whole transcriptome profiling is the sequencing-based 10x Genomics Visium set of assays.
Visium utilizes a technology where oligonucleotides are arrayed on a special glass slide in spatially arranged dots. Messenger RNA (mRNA) or probes targeting mRNA coming from tissue sections are transferred onto this Visium slide. Each spatial dot of oligonucleotides contains a specific barcode sequence identifying the spatial location, and the end of the oligo contains a complementary sequence to capture mRNA or probes coming from the tissue. Sequencing libraries are made from these spatially barcoded molecules, which are then read out on a NextGen sequencing platform such as an Illumina sequencer. The resulting sequencing data can identify the transcript identity and the spatial location from which it came from on the slide; this data, when overlayed onto the tissue section image, provides a whole transcriptome view of expression patterns in a spatial context.
Initial versions of the Visium technology required fresh frozen tissue sections (and later, an option to use FFPE tissue sections) to be placed specifically in a small window on to a relatively expensive Visium capture slide. This could be technically challenging, limited some of the eligible sample sources, and required optimized tissue permeabilization in the case of fresh frozen sections or tissue adherence optimization in the case of FFPE sections. Recently, the introduction of the 10x Genomics CytAssist platform has simplified the workflow by allowing sections to be cut onto normal glass slides. Following tissue preparation and imaging, a whole transcriptome probe set is then hybridized to transcripts in the tissue. Subsequently, the probes are transferred using the CytAssist instrument to the Visium spatially barcoded slide and the rest of the Visium workflow continues. Since adopting the CytAssist instrument, we have seen not only improvements in the workflow flexibility and sample accessibility, but also some improvements in the technical performance of the Visium assay.
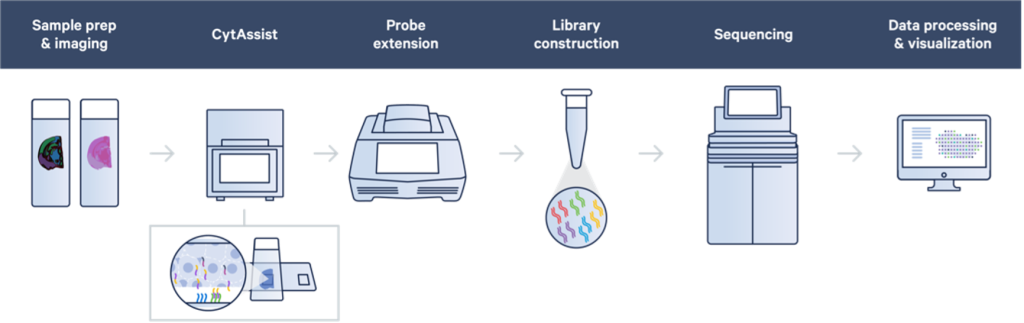
Visium FFPE v2 with CytAssist Workflow – Diagram from 10x Genomics, Inc
One current limitation of the existing version of the Visium platform is resolution of the resulting data. Spatially barcoded spots are 55 microns in diameter and are 100 microns apart from each other, center-to-center. Although there is considerable variation between tissues, in this format, the resulting data from Visium is often not at a single cell resolution. Spatially resolved data from Visium for each given barcode is typically an average expression profile of multiple cells. Computational methods to deconvolve mixed transcriptional profiles in spatial datasets exist, and these are often aided by matched single cell RNA-Seq data, but the performance of these tools may not resolve all caveats. Although a higher resolution version of Visium (Visium HD) has been announced by 10x Genomics, the exact date when this will be available is currently unknown.
Other spatial transcriptomic profiling methods and platforms exist, and in some cases these may help generate data that augments Visium data, or in some cases, may be better suited to answer specific biological questions. In this fast-paced spatial profiling technology landscape, it is recommended that investigators consult with some of the core facilities that support existing spatial assays and platforms, and who are testing emerging technologies. CCR’s Office of Science and Technology Resources helps coordinate these efforts and can also help advise on where CCR Investigators can find relevant support.
As with single cell sequencing experiments, the investment in these cutting-edge technologies must also include careful experimental design, optimized samples preparation, and downstream informatic analysis and interpretation of the complex data. Smaller pilot projects are highly recommended before planning Visium projects with larger sample sets.
If you are interested in seeking support on the 10x Genomics Visium platform, please reach out to CCR Single Cell and Spatial Core (CCR SCSC). Currently, the 10x Genomics Visium FFPE v2 with CytAssist-enabled workflow for human or mouse tissue is supported by CCR SCAF. Labs without their own histology capabilities often require assistance with the upstream sample processing required for Visium assays. The Molecular Histopathology Lab (MHL) in Frederick has provided CCR Investigators with expert upstream support for Visium workflows, including histology, sectioning and staining of tissue sections. In addition to working closely to test and optimize the Visium and other spatial assay workflows, MHL and SCAF are partnering to provide expert technical and efficient project support CCR Investigators.
For CCR Investigator labs performing Visium experiments in-house and have libraries to sequence, the Frederick Sequencing and Genomics Core (FSGC) in Frederick and the CCR Genomics Core have the necessary experience to sequencing these library types and assist with processing the resulting sequence data.
— Michael Kelly, Jatinder Singh and other members the CCR Single Cell Analysis Facility Team