Tips for jupyter lab
Learning Objectives
After this coding club session, the audience should
- Have an appreciation of Jupyter Lab as an all-in-one tool for documenting code, output, and analysis steps
- Have the foundational knowledge needed to start using Jupyter Lab
- Become familiar with resources for self and continual learning of Jupyter Lab
- Be acquainted with methods for sharing Jupyter Lab
Note that this class is not hands-on
Markdown editing tips for Jupyter Notebook
Jupyter Notebook utilizes markdown for text editing.
"Markdown is is a lightweight markup language for creating formatted text using a" -- https://en.wikipedia.org/wiki/Markdown
We can use different size headings to divide sections in a Jupyter Notebook. See https://www.markdownguide.org/basic-syntax/ for more details.
# Heading level 1
## Heading level 2
### Heading level 3
...
To make an unordered list in Jupyter notebook, use either - or *. Examples are below
Skills and knowledge needed for bioinformatics
- Biology
- Coding
- Math and statistics
Suggested computer skills for bioinfor
* Unix command line
* R
* Python
To make an ordered list, we can use numbers. As an example
Steps in bulk RNA sequencing analysis
1. Check sequencing data quality
2. Adapter and quality trimming
3. Re-check sequencing data quality after step 2
4. Align sequencing data to reference geneome or transcriptome
5. Perform post alignment quality check
6. Obtain gene expression counts table
7. Perform differential expression analysis
8. Perform pathway analysis
To make a nested list, see the following example where each sub-items are created by tab indentation on the next line.
- RNA sequencing aligners should be splice aware and include
- HISAT2
- STAR
- Differential expression analysis packages include
- DESeq2
- edgeR
- limma
Subsripts and superscripts
Subscripts
To create subscripts, enclose text in $ and then place the portion to be subscripted inside the curly brackets prepended by _.
$CO_{2}$
Superscripts
To create superscripts, enclose text in $ and then prepend ^ to the text that needs to be superscripted.
$x^2$
To bold text we wrap with "**" on both sides
**bold**
To italicize text wrap with "*" on both sides
*italicize*
To insert links
[](url)
To insert images

If we need to adjust image size, we will need to use HTML. See https://www.w3schools.com/tags/att_img_width.asp for inserting images and adjusting image size using HTML.
<img src="path to image" width="" height="" />
We can insert equations. Below is the equation for calculating z-score.
$$z-score=\frac{x-mean}{standard\ deviation}$$
$$x={-b \pm \sqrt{b^2-4ac} \over 2a}$$
$$sd=\sqrt{\frac{(x_i-mean)^2}{N}}$$
$$e=mc^2$$
We can insert tables into our Jupyter analysis documents. Below is a made-up table containing expression of two genes in control and drug groups.
| Gene | Control | Drug |
|---|---|---|
| BRCA1 | 200 | 400 |
| HGPRT | 500 | 150 |
Other tips for Jupyter
Start a Jupyter Lab session
Assuming Jupyter Lab has been installed on a personal computer, it can be started using the Mac OS Terminal or Windows Command Prompt by typing jupyter lab --no-browser, which will initiate the Jupyter Lab session and provide you with a local host link that can be inserted into a web browser, where you will be interfacing with Jupyter Lab.
To access the server, open this file in a browser:
file:///Users/wuz8/Library/Jupyter/runtime/jpserver-67498-open.html
Or copy and paste one of these URLs:
http://localhost:8889/lab?token=ea3018c348569081f64232675b04be67b4a012c0a0a33e76
or http://127.0.0.1:8889/lab?token=ea3018c348569081f64232675b04be67b4a012c0a0a33e76
To exit Jupyter Lab, remember to save your work and then go back to Mac OS Terminal or Windows Command Prompt and hit control-c. Select y when asked if you like to "Shutdown this Jupyter server"
Shutdown this Jupyter server (y/[n])?
Jupyter Lab Launcher
Upon starting Jupyter Lab, users will see a launcher that allows for selection of the tool that they like to use.
Notebook: Under the "Notebook" section, we can start a new notebook according to language needed for our data analysis. By default, Jupyter supports Python. Thus, if you just installed Jupyter then you will only be able to open a notebook that run Python. To run a notebook for R, you will need to install a R Jupyter Kernel (see https://cran.r-project.org/web/packages/IRkernel/readme/README.html. For more languages supported by Jupyter, see https://github.com/jupyter/jupyter/wiki/Jupyter-kernels.
"Kernels are programming language specific processes that run independently and interact with the Jupyter Applications and their user interfaces." -- [See Jupyter documentation on Kernels](https://docs.jupyter.org/en/latest/projects/kernels.html)`
Console: We can open open a console to run code and view output in a lanugage of our choice. We can't save work that was done in console.
Others: We can do the following
- Access a terminal
- Open text files
- Open scripts, depending on what language the user has installed
To check the available kernels, goto the Terminal (Mac) or Command Prompt (Windows) and type jupyter kernelspec list
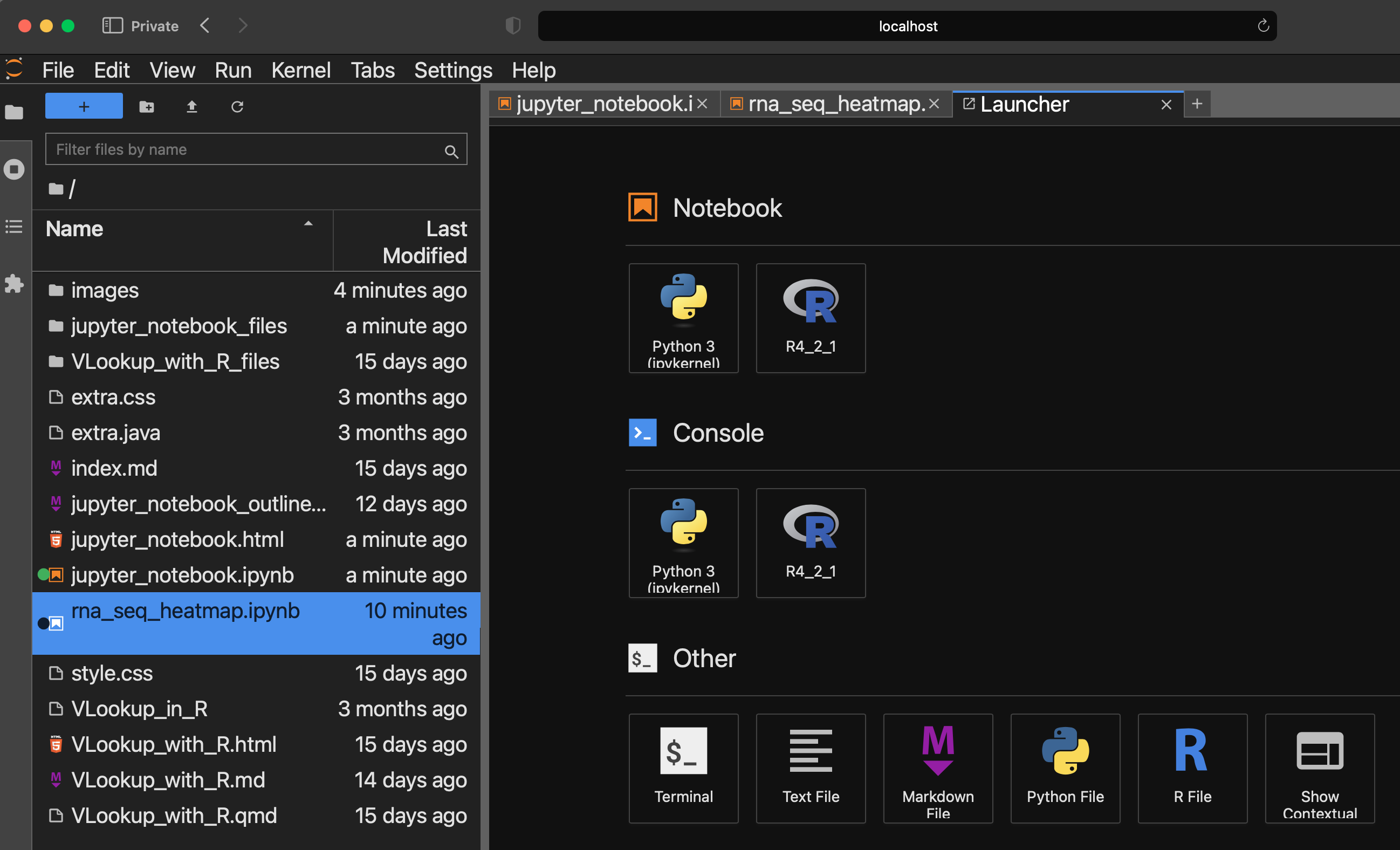
Jupyter Lab Explorer and Notebook Panels
The Jupyter Lab Interface is divided into an explorer panel where users can access files and a panel where users will work with notebooks. It is the notebooks where users work with code and document output as well as analysis steps. Multiple notebooks can be opened at the same time.
All notebooks created in Jupyter Lab have extension ipynb, which stands for Interactive Python Notebook.
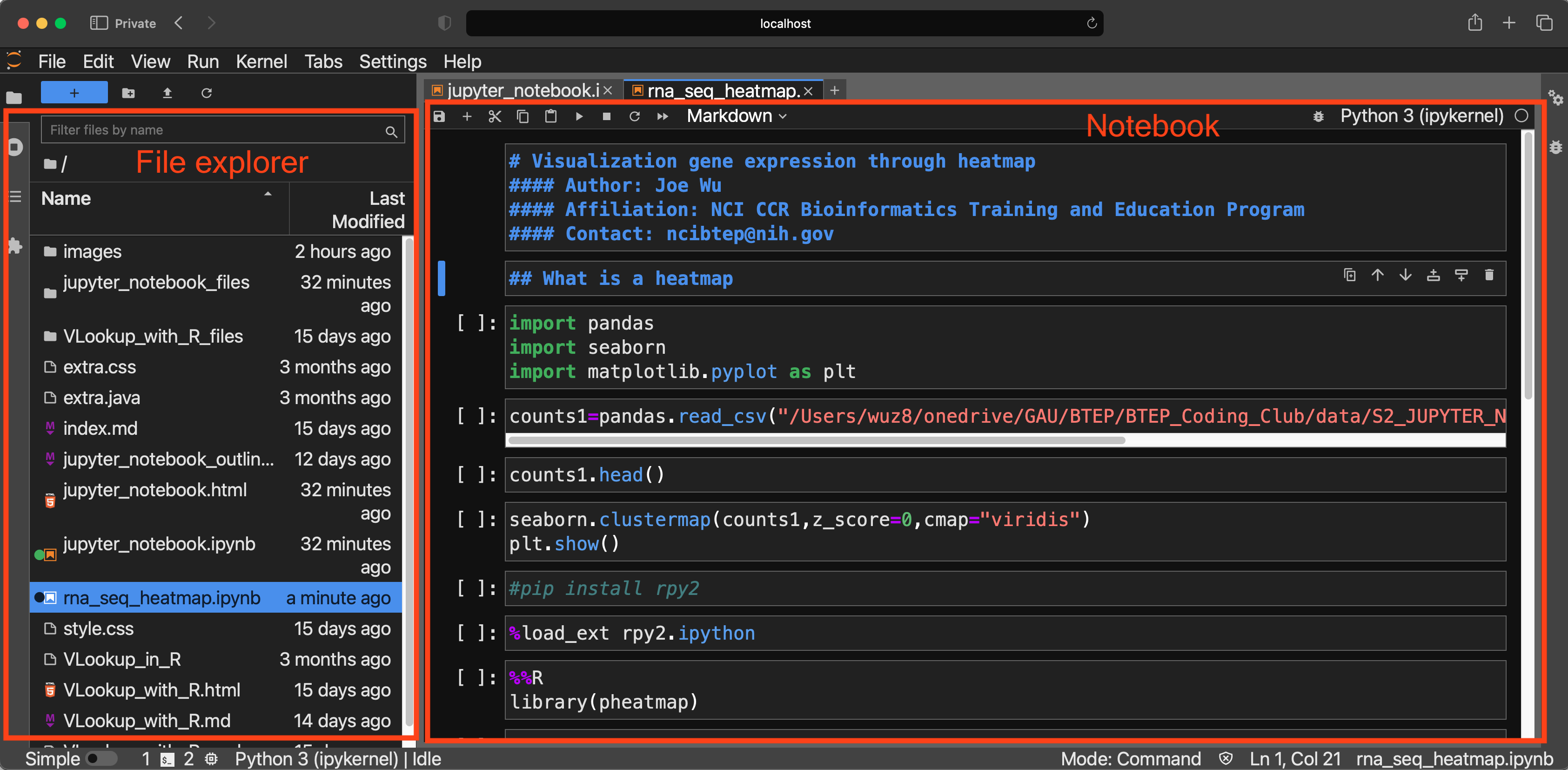
Jupyter Lab Menu Bar
The menu bar in Jupyter Lab contains useful menu. These include:
- File
- Open
- Save
- Export
- HTML
- Markdown
- Run
- Kernel for restarting a notebook and clearing output

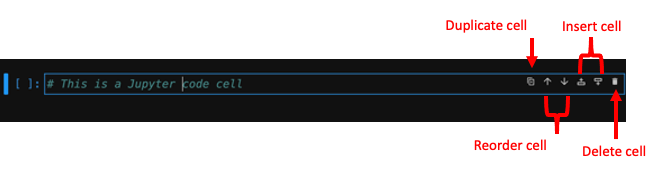
The Jupyter Notebook Cell
The basic working unit of a Jupyter Notebook is the cell, where users can enter code or formatted text.
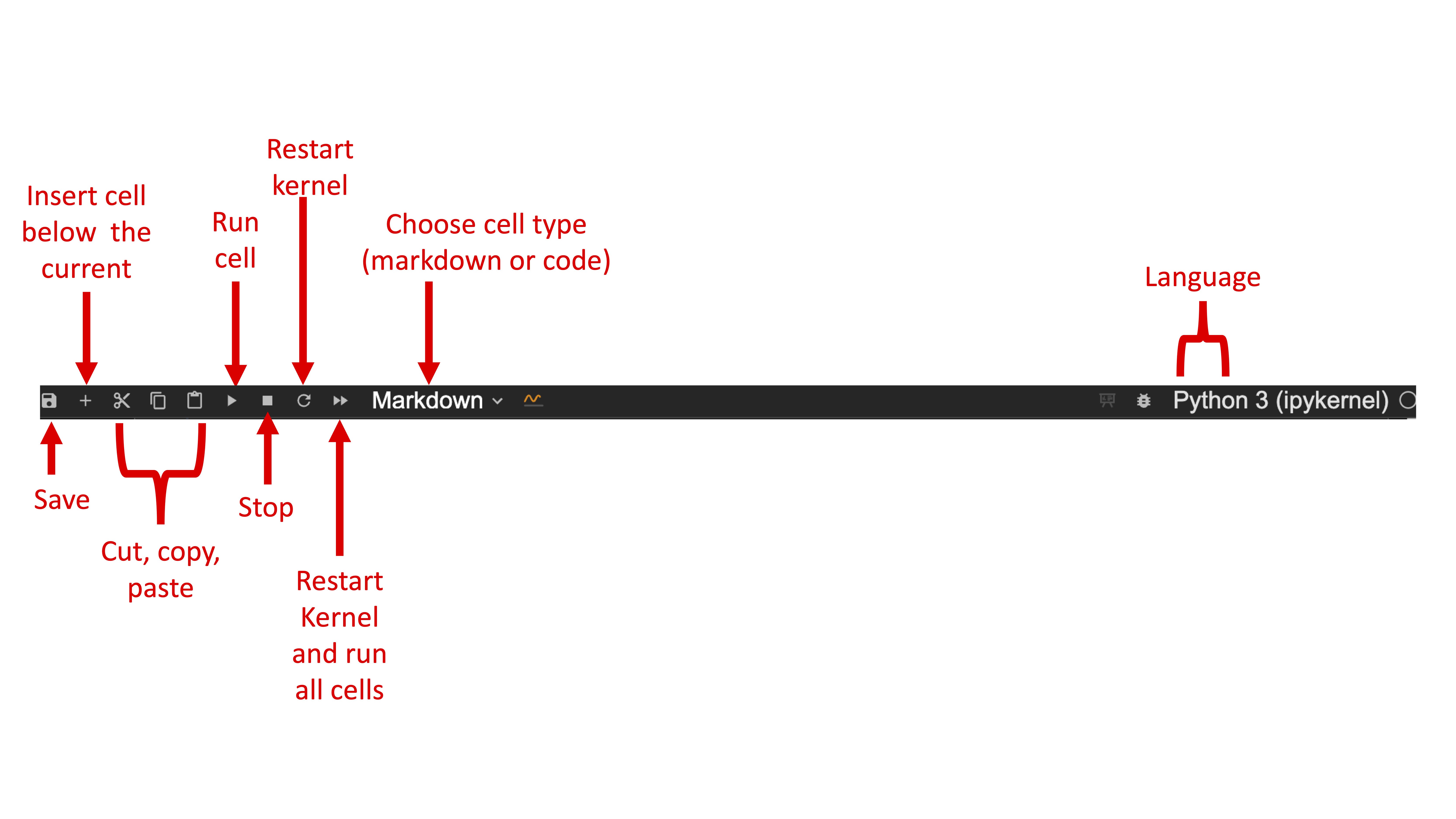
Jupyter Notebook Menu Bar
In the notebook panel of Jupyter Lab, there is a menu bar that allows users to save, cut/copy and paste and run cells. It also informs users which language the notebook is running. Importantly, the power of Jupyter lies in its ability to allow users write text as well insert relevant images and tables to more efficiently document their code and output. Thus, it is important for users to be able to choose whether markdown or code is included in a cell.
Accessing Jupyter Lab
On personal computer
- Miniconda
- Need to install Jupyter Note separately (see https://jupyter.org/install)
- Anaconda
On NIH High Performance Computing
- Biowulf (see Jupyter on Biowulf)
Note: to install software on your government furnished computer, submit a ticket with https://service.cancer.gov/ncisp
Exporting Jupyter Lab
We can use the "Save and Export Notebook As" in the File menu to export our notebook to different formats including HTML, PDF, and Markdown.
Alternatively, we can save using the command line. The command is jupyter nbconvert. We can use jupyter nbconvert --help to find out more about our export options. Like other Unix commands, we can append --help to read about Jupyter commands.
The syntax for exporting is:
jupyter nbconvert name_of_notebook.ipynb --to format
Where format can include but not limited to HTML, PDF, and Markdown.
Ways to share Jupyter Notebook
- Github (https://github.com)
- Binder (https://mybinder.org) - users can run code
- See https://mybinder.org/v2/gh/drice-codeathons/workshop-ncbi-data-with-python/main?filepath=notebooks%2Fworkshop.py for an example of a Jupyter Notebook shared on Binder
Useful Jupyter Extensions
- Ipywidgets - to create interactive notebook elements
- MyST - provides more text editing features including admonitions such as notes, warnings, and cautions
Jupyter Help Resources
- Jupyter Project Documentation
- Jupyter Notebook Readthedocs
- Click on Help in the Jupyter Lab menu bar to pull help resources
- Markdown tutorial