Data Analysis
Here are a pair of examples of RNASEQ complete workflows
RNASEQ Pipeline from NCI CCBR
https://github.com/CCBR/Pipeliner/blob/master/RNASeqDocumentation.pdf

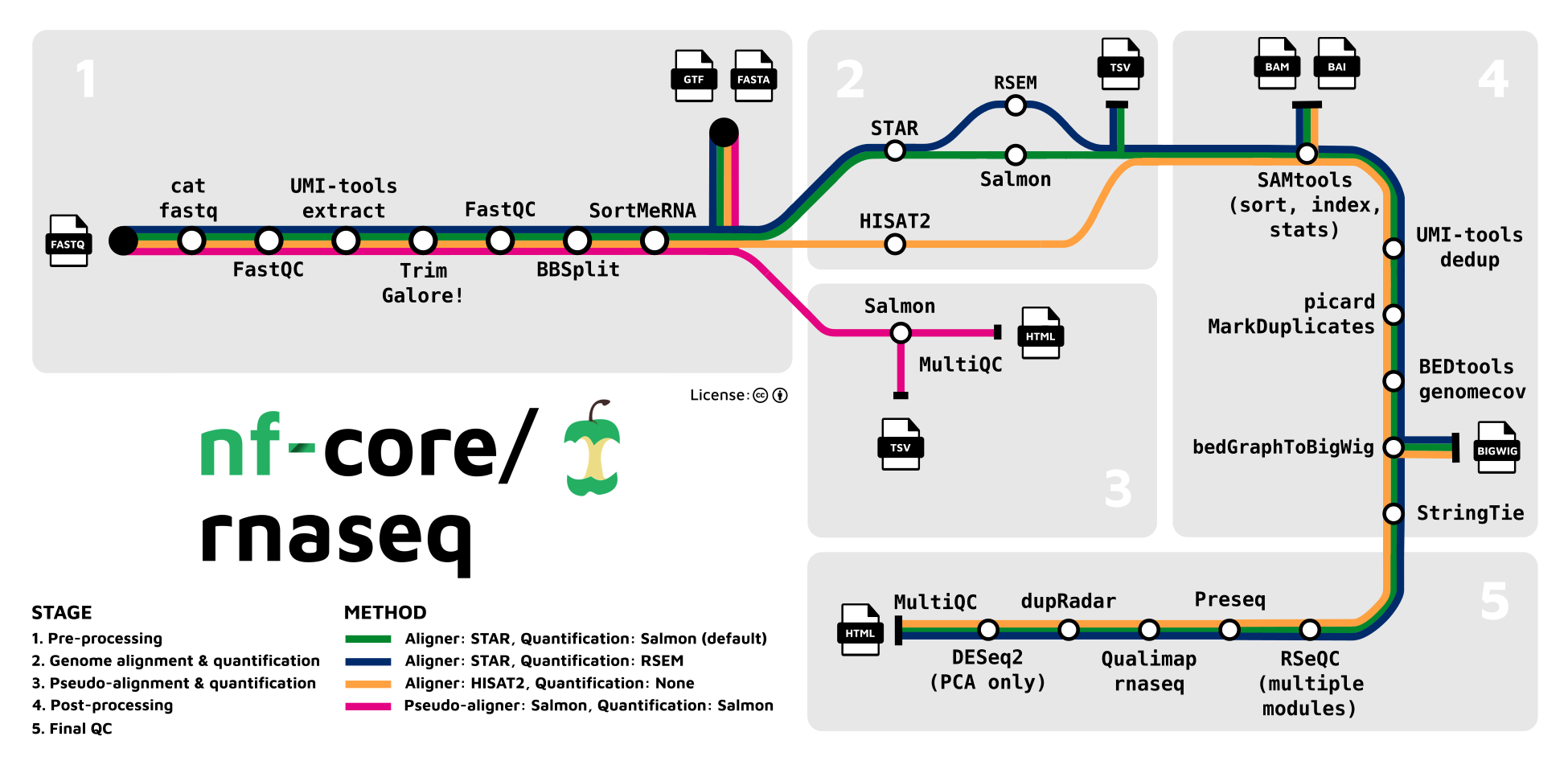
RNASEQ Nextflow Pipeline from nf-core
https://nf-co.re/rnaseq
Data Quality Assessment (QC)
Basic RNASEQ Quality Control (QC) examines the technical characteristics of the data produced by the sequencer. (It tells us nothing about whether the experiment worked. It answer the questions:
- Is the data of sufficiently high quality to be analyzed?
- Are there technical artifacts?
-
Are there poor quality samples?
-
It evaluate the following features
- Overall sequencing quality scores and distributions
- GC content distribution
- Presence of adapter or contamination
- Sequence duplication levels
Data should be filtered, trimmed, or rejected as appropriate
Sequencing cores generally provide some/all of this analysis
Examples of some of the quantities measured in basic QC
By the program FastQC
https://www.bioinformatics.babraham.ac.uk/projects/fastqc/good_sequence_short_fastqc.html
 GOOD
GOOD
 BAD
BAD
By the program MultiQC
https://multiqc.info/examples/rna-seq/multiqc_report.html

Raw Sequence Cleanup
Trim and/or filter sequence to remove sequencing primers/adaptor and poor quality reads. Example programs:
- FASTX-Toolkit is a collection of command line tools for Short-Reads FASTA/ FASTQ files preprocessing.
- SeqKit is an ultrafast comprehensive toolkit for FASTA/Q processing.
- Trimmomatic is a fast, multithreaded command line tool that can be used to trim and crop Illumina (FASTQ) data as well as to remove adapters.
- TrimGalore is a wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files, with some extra functionality for MspI-digested RRBS-type (Reduced Representation Bisufite-Seq) libraries.
- Cutadapt finds and removes adapter sequences, primers, poly-A tails and other types of unwanted sequence from your high-throughput sequencing reads.